
Projects
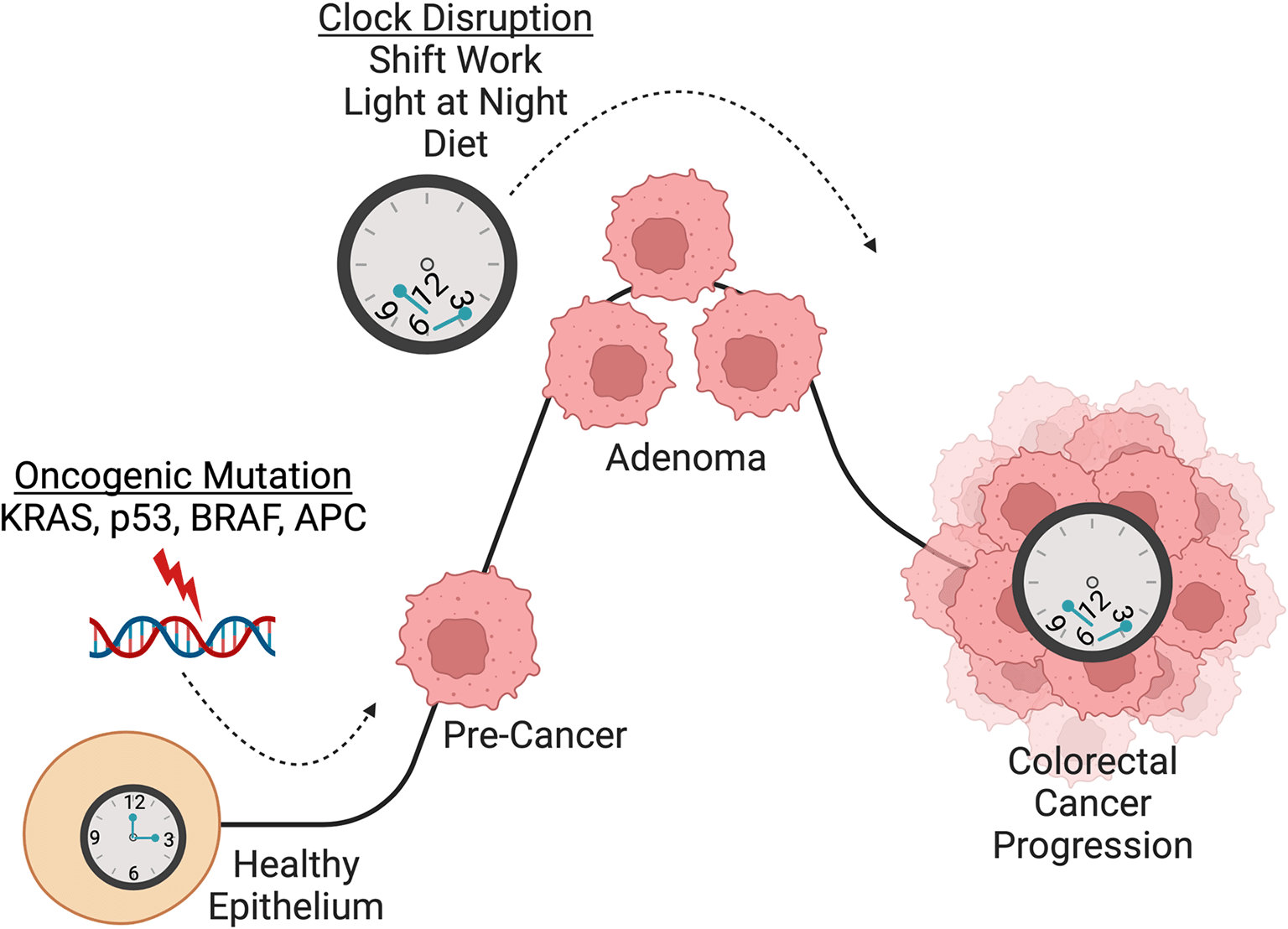
image from DOI (Fortin, 2023)
01
“Unraveling rhythmic crosstalk underlying CRC progression using single cell profiling and next-generation organoids” (Postdoc)
Circadian disruption, perturbation of endogenous circadian clocks, has been classified as a probable carcinogen and, due to artificial lighting, shiftwork, and international travel, is common in our modern 24/7 society. Increasing evidence points towards the mechanistic involvement of endogenous circadian clocks in CRC development and progression, providing unexplored opportunities to better understand and treat this disease. Therefore, this project aims to unravel the role of circadian clocks in CRC progression using single cell profiling and next-generation organoids. We will study colorectal tumors on single cell level to identify rhythmically regulated, multicellular interaction networks underlying CRC progression. We will develop next-generation organoids, i.e. 3D co-cultures of patient derived epithelial organoids with other components of the tumor microenvironment (TME), to model such interactions between immune, stromal, and malignant cells. Through intervention studies, we will ascertain the mechanistic connection between endogenous circadian clocks and CRC progression.
02
“Defining the physiological role of TGF-beta signaling for intercellular clock coupling within peripheral tissues” (Postdoc)
We previously identified the TGF-beta (TGFb) signaling pathway as a candidate pathway for intercellular oscillator coupling within peripheral clock networks. Based on theoretical predictions and studies on the central pacemaker clock, oscillator coupling governs how tissue clocks respond to incoming zeitgeber signals, to which environmental cycles they can entrain, as well as how stable their rhythms are under free-running conditions. We predict that disruption of the TGFb pathway within organ clocks (e.g. the liver), and thus perturbed intercellular coupling between cellular oscillators, will render tissue rhythms more susceptible to perturbation by external zeitgebers. As a consequence rhythmic output functions of the affected organ are expected to be altered in phase and/or amplitude and entrainment to environmental cycles should be changed. Ultimately, this may contribute to circadian disruption and promote associated pathologies.

image from PMID (Vetter, 2018)

03
“Unraveling mechanisms of intercellular communication between peripheral circadian clocks” (PhD thesis)
The molecular machinery that generates circadian rhythms can be found in virtually any cell type. Coupling between cell-autonomous circadian oscillators is crucial to prevent desynchronization and damping of circadian tissue rhythms. Such tissue rhythms are essential for the temporal coordination of organ functions, as well as their adaptation to cyclic environmental changes. While mechanisms of intercellular coupling within the suprachiasmatic nucleus, the mammalian pacemaker clock in the brain, are well described, we lack a detailed understanding of peripheral oscillator coupling. Because in vitro models of peripheral clocks show characteristics of weak coupling, we aimed to identified signaling pathways that mediate intercellular communication and synchronization of cell-autonomous oscillations.
04
“Studying interactions between RNA binding proteins and the polyadenylation factor CPSF6” (MSc thesis)
A genome-wide RNAi screen in U2OS cells identified CPSF6, a key regulator of 3'end cleavage and polyadenylation, as important regulator of circadian period and temperature compensation. Knock-down of this gene induces a global upstream shift of PAS usage and shortening of 3'UTRs, which might interfere with regulation of mRNAs by RNA binding proteins (RBP). We hypothesized that such (post)-transcriptional regulatory mechanisms govern temperature compensation of circadian clocks and aimed to identify interaction partners of CPSF6 that are involved in PAS usage dependent regulation of circadian dynamics.
